Automated processing pipelines using EEGLAB
You do not need to write a script to process all datasets simultaneously in a STUDY. This video describes how to perform batch processing from the EEGLAB graphic interface. In this section, we run a similar pipeline using a script.
Table of contents
Creating a STUDY or import from BIDS
Download the data from https://openneuro.org/datasets/ds003061/ and go to this folder, then run the script in the next section.
Alternatively, use one of the available EEGLAB studies. Note that some of these studies already have their data preprocessed and may not be suitable for automated processing.
Running an ERP pipeline
The pipeline below takes the raw data from all subjects, clean the data, extracts epochs of interest, and plots the grand average ERP.
First, the data is imported. Then it is cleaned with clean_rawdata (default parameters are used here but may need adjustment based on data quality). ICA is then run on all datasets, and ICLabel is used to flag bad ICA components.
Then oddball and standard stimuli epochs are extracted, the baseline is removed (from -1000 to 0 millisecond before the stimulus onset), and ERPs are computed and plotted. If you want to run this pipeline on your own EEGLAB study, then you will need to modify the line for extracting data epochs (as the name of your events will likely differ).
% check folder
eeglab;
if ~exist('task-P300_events.json', 'file')
error('Download the data from https://openneuro.org/datasets/ds003061/ and go to the downloaded folder');
else
filepath = fileparts(which('task-P300_events.json'));
end
% import data
pop_editoptions( 'option_storedisk', 1); % only one dataset in memory at a time
[STUDY, ALLEEG] = pop_importbids(filepath, 'studyName','Oddball', 'subjects', [1:2]); % when using all subjects, one subject is truncated and cause the pipeline to return an error
% remove non-ALLEEG channels (it is also possible to process ALLEEG data with non-ALLEEG data
ALLEEG = pop_select( ALLEEG,'rmchannel',{'EXG1','EXG2','EXG3','EXG4','EXG5','EXG6','EXG7','EXG8', 'GSR1', 'GSR2', 'Erg1', 'Erg2', 'Resp', 'Plet', 'Temp'});
% compute average reference
ALLEEG = pop_reref( ALLEEG, []);
% clean data using the clean_rawdata plugin
ALLEEG = pop_clean_rawdata( ALLEEG,'FlatlineCriterion',5,'ChannelCriterion',0.87, ...
'LineNoiseCriterion',4,'Highpass',[0.25 0.75] ,'BurstCriterion',20, ...
'WindowCriterion',0.25,'BurstRejection','on','Distance','Euclidian', ...
'WindowCriterionTolerances',[-Inf 7] ,'fusechanrej',1);
% recompute average reference
ALLEEG = pop_reref( ALLEEG,[]);
% run ICA reducing the dimension by 1 to account for average reference
plugin_askinstall('picard', 'picard', 1); % install Picard plugin
ALLEEG = pop_runica(ALLEEG, 'icatype','picard','concatcond','on','options',{'pca',-1});
% run ICLabel and flag artifactual components
ALLEEG = pop_iclabel(ALLEEG, 'default');
ALLEEG = pop_icflag( ALLEEG,[NaN NaN;0.9 1;0.9 1;NaN NaN;NaN NaN;NaN NaN;NaN NaN]);
% Optional: remove flagged ICA components (otherwise done at the STUDY level), then recompute the
% average reference using the Huber method interpolating missing channels (and removing them again
% after average reference). See tutorial section 5.b.
if 0
ALLEEG = pop_subcomp(ALLEEG, []);
ALLEEG = pop_reref( ALLEEG, [], 'huber', 25, 'interpchan',[], 'refica', 'remove');
end
% extract data epochs
ALLEEG = pop_epoch( ALLEEG,{'oddball_with_reponse','standard'},[-1 2] ,'epochinfo','yes');
ALLEEG = eeg_checkset( ALLEEG );
ALLEEG = pop_rmbase( ALLEEG,[-1000 0] ,[]);
% create STUDY design
STUDY = std_maketrialinfo(STUDY, ALLEEG);
STUDY = std_makedesign(STUDY, ALLEEG, 1, 'name','STUDY.design 1','delfiles','off', ...
'defaultdesign','off','variable1','type','values1',{'oddball_with_reponse','standard'},...
'vartype1','categorical','subjselect', STUDY.subject);
% precompute ERPs at the STUDY level
[STUDY, ALLEEG] = std_precomp(STUDY, ALLEEG, {},'savetrials','on','rmicacomps','on','interp','on','recompute','on','erp','on');
% plot ERPS
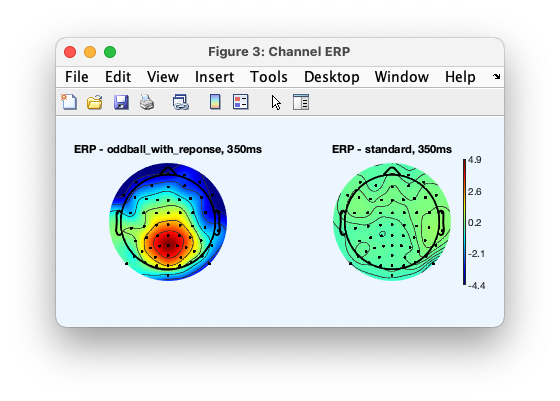
STUDY = pop_erpparams(STUDY, 'topotime',350);
chanlocs = eeg_mergelocs(ALLEEG.chanlocs); % get all channels from all datasets
STUDY = std_erpplot(STUDY,ALLEEG,'channels', {chanlocs.labels}, 'design', 1);
% revert default option
pop_editoptions( 'option_storedisk', 0);
A figure similar to the one below will be plotted. The figure may differ as some of the artifact and rejection steps above involve choosing data randomly. To make the pipeline reproducible, add “rng(1)” at the beginning of the script above. note that the script above only process the first two participants. Update the call to pop_importbids() to process all participants.
Running a spectral pipeline
The pipeline below takes the raw data from all subjects, clean the data, extracts epochs of interest, and plots the spectrum to compare conditions. The first part is identical to the ERP script above. The end of the script computes the spectrum. Note that if you have continuous data, you need not extract epochs. We extracted epochs in this case since we wanted to reuse the same dataset as above.
% check folder
eeglab;
if ~exist('task-P300_events.json', 'file')
error('Download the data from https://openneuro.org/datasets/ds003061/ and go to the downloaded folder');
else
filepath = fileparts(which('task-P300_events.json'));
end
% import data
pop_editoptions( 'option_storedisk', 1); % only one dataset in memory at a time
[STUDY, ALLEEG] = pop_importbids(filepath, 'studyName','Oddball', 'subjects', [1:2]); % when using all subjects, one subject is truncated and cause the pipeline to return an error
% remove non-ALLEEG channels (it is also possible to process ALLEEG data with non-ALLEEG data
ALLEEG = pop_select( ALLEEG,'rmchannel',{'EXG1','EXG2','EXG3','EXG4','EXG5','EXG6','EXG7','EXG8', 'GSR1', 'GSR2', 'Erg1', 'Erg2', 'Resp', 'Plet', 'Temp'});
% compute average reference
ALLEEG = pop_reref( ALLEEG, []);
% clean data using the clean_rawdata plugin
ALLEEG = pop_clean_rawdata( ALLEEG,'FlatlineCriterion',5,'ChannelCriterion',0.87, ...
'LineNoiseCriterion',4,'Highpass',[0.25 0.75] ,'BurstCriterion',20, ...
'WindowCriterion',0.25,'BurstRejection','on','Distance','Euclidian', ...
'WindowCriterionTolerances',[-Inf 7] ,'fusechanrej',1);
% recompute average reference interpolating missing channels (and removing
% them again after average reference - STUDY functions handle them automatically)
ALLEEG = pop_reref( ALLEEG, []);
% run ICA reducing the dimension by 1 to account for average reference
plugin_askinstall('picard', 'picard', 1); % install Picard plugin
ALLEEG = pop_runica(ALLEEG, 'icatype','picard','concatcond','on','options',{'pca',-1});
% run ICLabel and flag artifactual components
ALLEEG = pop_iclabel(ALLEEG, 'default');
ALLEEG = pop_icflag( ALLEEG,[NaN NaN;0.9 1;0.9 1;NaN NaN;NaN NaN;NaN NaN;NaN NaN]);
% Optional: remove flagged ICA components (otherwise done at the STUDY level), then recompute the
% average reference using the Huber method interpolating missing channels (and removing them again
% after average reference). See tutorial section 5.b.
if 0
ALLEEG = pop_subcomp(ALLEEG, []);
ALLEEG = pop_reref( ALLEEG, [], 'huber', 25, 'interpchan',[], 'refica', 'remove');
end
% extract data epochs
% this is not necessary if you have resting state data or eyes open
% eyes closed data, you need to define the design in the STUDY
ALLEEG = pop_epoch( ALLEEG,{'oddball_with_reponse','standard'},[-1 2] ,'epochinfo','yes');
ALLEEG = eeg_checkset( ALLEEG );
% create STUDY design
STUDY = std_maketrialinfo(STUDY, ALLEEG);
STUDY = std_makedesign(STUDY, ALLEEG, 1, 'name','STUDY.design 1','delfiles','off', ...
'defaultdesign','off','variable1','type','values1',{'oddball_with_reponse','standard'},...
'vartype1','categorical','subjselect', STUDY.subject);
% precompute ERPs at the STUDY level
[STUDY, ALLEEG] = std_precomp(STUDY, ALLEEG, {},'savetrials','on','rmicacomps','on','interp','on','recompute','on','spec','on');
% plot ERPS
STUDY = pop_specparams(STUDY, 'topofreq',10);
chanlocs = eeg_mergelocs(ALLEEG.chanlocs); % get all channels from all datasets
STUDY = std_specplot(STUDY,ALLEEG,'channels', {chanlocs.labels}, 'design', 1, 'ylim', [40 55]););
% revert default option
pop_editoptions( 'option_storedisk', 0);
A plot similar to the following one will appear. It might be slightly different as the options to remove the artifacts above have been changed.

Optimizing the pipeline for your data
Filtering
You might want to apply a different filter than the filter applied by the clean_rawdata plugin (which is an elliptic filter). For example, to apply a standard FIR filter, you would need to replace the call to the clean_artifacts function by:
EEG = pop_eegfiltnew( EEG,'locutoff',0.5);
EEG = clean_artifacts( EEG,'FlatlineCriterion',5,'ChannelCriterion',0.8, ...
'LineNoiseCriterion',4,'Highpass','off' ,'BurstCriterion',20, ...
'WindowCriterion',0.25,'BurstRejection','on','Distance','Euclidian', ...
'WindowCriterionTolerances',[-Inf 7] ,'fusechanrej',1);
Note that when calling the function to clean artifacts from the clean_rawdata plugin, the highpass argument is set to off to disable filtering.
Why do we reference twice?
Artifact cleaning using clean_rawdata usually works better on averaged reference data. We have made this observation when processing data, although there is no published article on this topic. After bad channels have been removed, then we need to compute the average reference again. The second average reference computation undoes the first one, as explained on this page.
Automated cleaning parameter
The call to clean_artifacts uses the default EEGLAB parameters. However, these are not always optimal. In particular, ‘ChannelCriterion’ may be modified to reject more or fewer channels. ‘BurstCriterion’ is another important parameter. Increase it to 40, for example (or some people recommend 100) if you feel too many data regions are rejected. More information is available on the plugin wiki page.
Finding optimal parameters for cleaning your data is essential to designing your pipeline. It would be best if you experimented with a couple of subjects. Also, once you run your pipeline on all subjects, you should check how much data was removed for each subject. Sometimes, 80% of the data is removed for some subjects, which is not acceptable.
An alternative solution for cleaning data is also to run data cleaning multiple times. Your run artifact rejection once to remove bad channels and large artifacts. Then after running ICA, you can run it again to remove smaller artifacts. The advantage of this approach is that the first data cleaning will not remove eye blinks (which ICA can subtract from your data allowing you to keep these regions of data). An example of this approach is shown in this section of the tutorial.
Which ICA to use
The script above uses runica (Infomax), which is the default in EEGLAB. Other popular choices which require installing the relevant plugins are Amica, Picard, and FastICA. We compared different ICA solutions in this paper. In short, there is no ideal algorithm:
- Runica has been used the most. It is robust but slow.
- picard optimizes the same objective function as runica. It converges faster and with lower residuals. It was also designed by one of the ICA pioneers, Jean-Francois Cardoso. Nevertheless, it is still a new algorithm (as of 2022) that has not been thoroughly compared with others on EEG data.
- FastICA is an ICA algorithm widely used on EEG data. The author of the algorithm advises using the symmetric approach instead of the iterative one (which is the default).
- Amica is the best ICA algorithm based on our comparison. Nevertheless, it is slow. Also, it may only be applied to single EEG datasets.
What algorithm for automated ICA artifact rejection?
We used ICLabel in the script above. They are others. For example MARA is another popular EEGLAB plugin to detect artifactual ICA components.
For ICLabel, you may set the threshold to detect artifactual components. In the script above, we set the threshold to 90% for the likelihood to be an eye movement artifact (blink or lateral eye movement) and 90% for the likelihood to be a muscle. This is quite conservative and will only reject 1 to 5 components per subject. Some researchers are less conservative and would set the threshold lower. The ICLabel page contains more information on this subject.
Finding dipoles for ICA component and ICA component clustering
Finding dipoles for ICA component and ICA component clustering may also be done at the STUDY level, for example, using the small snippet of code below.
% find dipoles for all ICA components of all subjects
dipfitPath = fileparts(which('pop_dipfit_settings.m'));
EEG = pop_dipfit_settings( EEG,'hdmfile', fullfile(dipfitPath, 'standard_BEM', 'standard_vol.mat'), ...
'coordformat','MNI','mrifile',fullfile(dipfitPath, 'standard_BEM', 'standard_mri.mat'), ...
'chanfile',fullfile(dipfitPath, 'standard_BEM','elec','standard_1005.elc'), ...
'coord_transform',[-4.8299e-05 1.4553e-05 -0.00010483 2.9747e-06 5.8989e-06 -1.5708 1 1 1] );
EEG = pop_multifit( EEG,[],'threshold',100,'plotopt',{'normlen','on'});
% cluster dipoles which are close to each other and plot one cluster
[STUDY ALLEEG] = std_preclust(STUDY, ALLEEG, 1,{'dipoles','weight',1});
[STUDY] = pop_clust(STUDY, ALLEEG, 'algorithm','Affinity Propagation');
STUDY = std_dipplot(STUDY,ALLEEG,'clusters',2, 'design', 1);
For more information, see the ICA clustering section of the tutorial.
More advanced pipelines
- What if I want to plot the spectrum instead of ERPs? The example above was for ERPs. However, it is easy to plot other measures as described in this page.
- Can I run statistics in my pipeline? Yes, you may plot ERPs and other measures and compute statistically significant regions as the tutorial explains in another section. You may also write custom code to generate figures for your papers.
- Can I use LIMO in my pipeline? Yes, of course. See this paper and the LIMO wiki for reference.
Other EEGLAB pipelines
Below are other EEGLAB pipelines.
- The PREP pipeline is an EEGLAB plugin. It is an outdated pipeline (as of 2022) because automated artifact detection is suboptimal, but it was a relevant pipeline from 2014 to 2020, and there is nothing fundamentally wrong with it.
- Makoto’s processing pipeline is another relevant reference. It is not recommended for beginners. Nevertheless, it contains important information about EEG processing, and it is a worthwhile read.
- Danielle Gruber’s pipeline. Danielle Gruber is an EEGLAB user who shared her pipeline. It is long and detailed and also outdated (as of 2022). We did not spot any fundamental errors in the pipeline, though. It is a single-subject pipeline.
- The BIDS data script is part of this tutorial, and it is a similar pipeline (although more complex) compared to the one presented on this page.
- The HAPPE pipeline. We have not evaluated this EEGLAB-based pipeline but it is a popular one.
- EPOC This project provides scripts to easily get into a standardized EEG analysis using EEGLAB. It is published in Frontiers in Neuroscience.
- Apice preprocessing pipeline for infant data.
- GUI to build EEGLAB-based pipelines EEG Signal Cleaning Pipeline Management.
- RELAX a fully automated EEGLAB pre-processing plugin using ICA and ICLabel.
Below is also a repository containing optimized EEGLAB, FieldTrip, Brainstorm, and MNE pipelines. We scanned parameters to find the optimal artifact rejection thresholds for each software package. We will let you guess which pipeline performed best 😊.
https://github.com/sccn/eeg_pipelines