Importing BIDS data
The Brain Imaging Data Structure (BIDS) data standards is a community standard for organizing, describing, and annotating collections of neuro-imaging datasets. A magnetoencephalography (MEG) data extension has been developed and, in 2019, another electrophysiology data extension for EEG and intracranial EEG (iEEG) data. EEGLAB allows importing and exporting EEG data to the BIDS format using the EEG-BIDS EEGLAB plugin.
Table of contents
Downloading a BIDS dataset
On openneuro.org, search for the term “EEG.” OpenNeuro will show all the EEG datasets. The quality of these datasets may vary, as shown in this 2021 article. We advise you to download this specific dataset, as shown below.
To download, create an empty folder on your computer, then press the download button and select this folder. Note that the web download interfaces sometimes fail (depending on the speed of your connection).
The alternative is either to use command-line programs openneuro, aws, or datalad. We have used the three of them with various degrees of success. Aws is somewhat simpler because it only requires one install. datalad requires installing other modules such as git and git-annex. The openneuro command is the only one that allows you not only to download but also upload a dataset back to OpenNeuro, and we recommend using it (uploading a dataset using the OpenNeuro web interface almost always systematically fails).
Importing the BIDS dataset as an EEGLAB study
Start EEGLAB, then install the bids-matlab-io EEGLAB plugin from the EEGLAB plugin manager using the File → Manage EEGLAB extensions menu item.
The select menu item File → BIDS tools → Import BIDS folder to STUDY and select the BIDS folder you have created. The following window pops up.
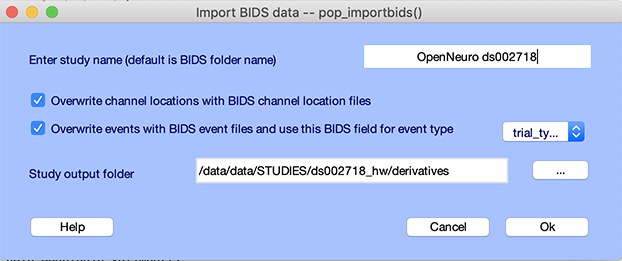
Raw EEG data files often define channel labels. However, BIDS also defines channel labels and channel locations in dedicated event files. By pressing the second checkbox, users may choose to use the channel label and location information contained in the BIDS channel definition files.
Raw EEG data file often has events. However, BIDS also define events in dedicated event files. Sometimes the BIDS event files contain more information than the raw EEG data file. In that case, users may choose to overwrite raw EEG data events with the event information contained in the BIDS event files. When BIDS events are selected, you may choose the BIDS event column to import, which may be “value” or “trial_type.” To choose which column to import, we recommend you look at one of the BIDS event files. In this case, we select trial_type.
Finally, users may select an output folder for storing their EEGLAB STUDY. If a folder is not selected, EEGLAB will store STUDY files ‘‘in place’’ which means in the BIDS folder structure - resulting in the BIDS folder becoming non-BIDS compliant and failing to pass BIDS validation because of the additional EEGLAB files.
Press Ok when done.
Processing BIDS EEGLAB study
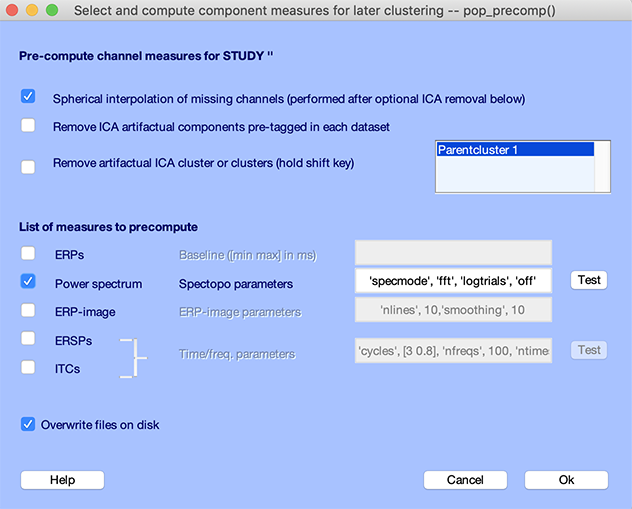
To illustrate that this EEGLAB STUDY is functional, we will simply plot the spectrum since we may do this without pre-processing. Select the Study → Precompute channel measures menu item, and check the Power spectrum checkbox, as shown below, then press Ok.
Then use menu item Study → Plot channel measures and, on the right column, press the Plot spectra button. The following plot pops up, showing the spectrum for all subjects.
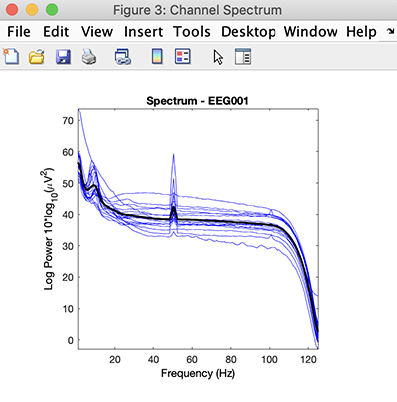
We can clearly see the alpha peak at about 10 Hz for most subjects (with one subject being an outlier, likely due to the presence of artifacts in his/her data).
You may follow the batch processing tutorial to pre-process all datasets simultaneously from the EEGLAB GUI, and perform standard EEGLAB group analyses.
You may also look at the BIDS script processing tutorial, which processes the same dataset from the MATLAB command line.