After editing the STUDY for group, we have the same Beta files as before, but also txt files split per group which makes file selection easier.
From the 2nd level GUI (figure 9), after creating/selecting a working directory and have selected your group level channel location file (likely in the /derivative root folder) select ANOVA, Repeated Measures ANOVA, Full scalp analysis, enter 2 groups, the finally select to use betas ; picking up iteratively list of beta files per group (figure 36). As before you also have to enter factors (here 3) and indicate the relevant beta parameters (here [1:3]).
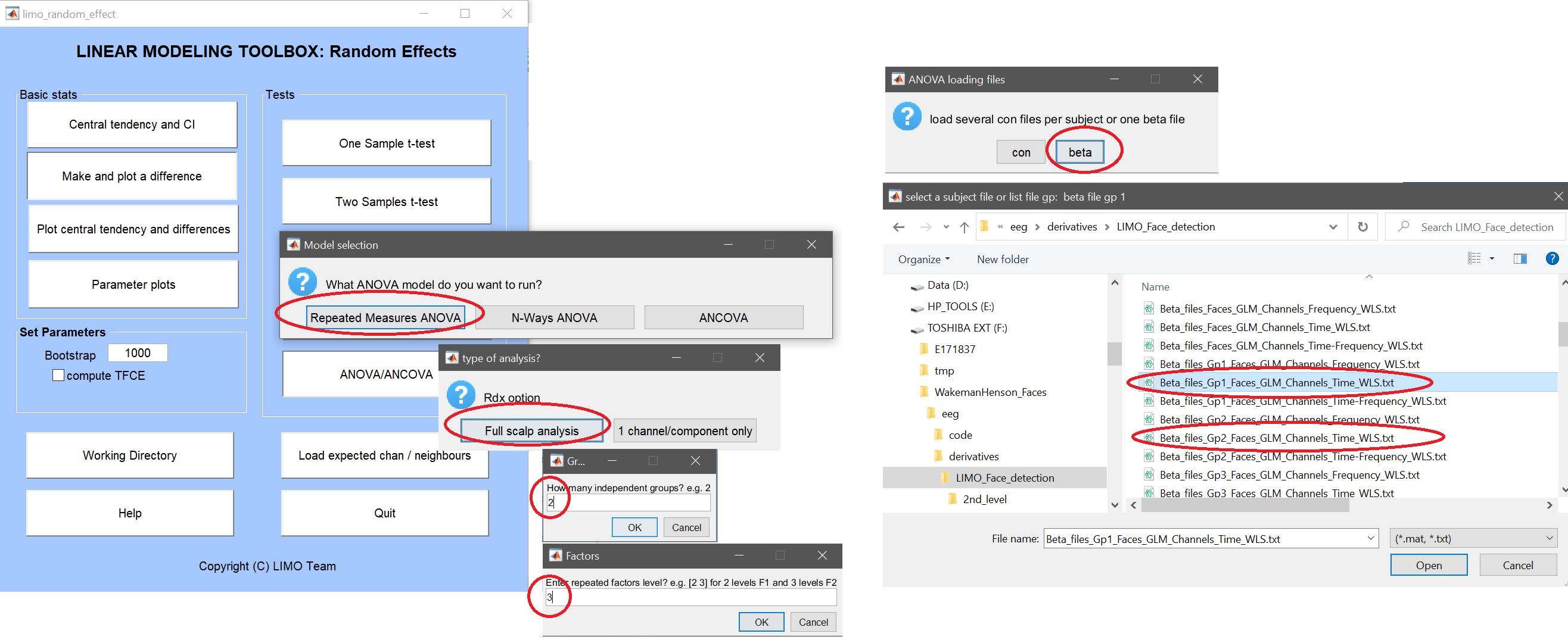 Figure 36. Gp * Repeated measure ANOVA
Figure 36. Gp * Repeated measure ANOVA
These steps can be executed in command line as:
mkdir('Gp-Conditions_ANOVA'); cd('Gp-Conditions_ANOVA');
chanlocs = [STUDY.filepath filesep 'limo_gp_level_chanlocs.mat'];
Files = cell(2,1); % groups in rows, repeated measures in columns
Files{1} = fullfile(STUDY.filepath,['LIMO_' STUDY.filename(1:end-6)],'Beta_files_Gp1_ANOVA_Faces_GLM_Channels_Time_WLS.txt');
Files{2} = fullfile(STUDY.filepath,['LIMO_' STUDY.filename(1:end-6)],'Beta_files_Gp2_ANOVA_Faces_GLM_Channels_Time_WLS.txt');
LIMOPath = limo_random_select('Repeated Measures ANOVA',chanlocs,'LIMOfiles',Files,...
'analysis_type','Full scalp analysis','parameters',{[1 2 3];[1 2 3]},...
'factor names',{'face'},'type','Channels','nboot',1000,'tfce',0,'skip design check','yes');
Output files are now the main effect of faces, the main effect of group and the interaction gp*faces. Results for the main effect are similar to the one-way ANOVA with faces as the only factor, but here we can test for the interaction group * faces as well (figure 37 for ERP, Spectrum and ERSP).
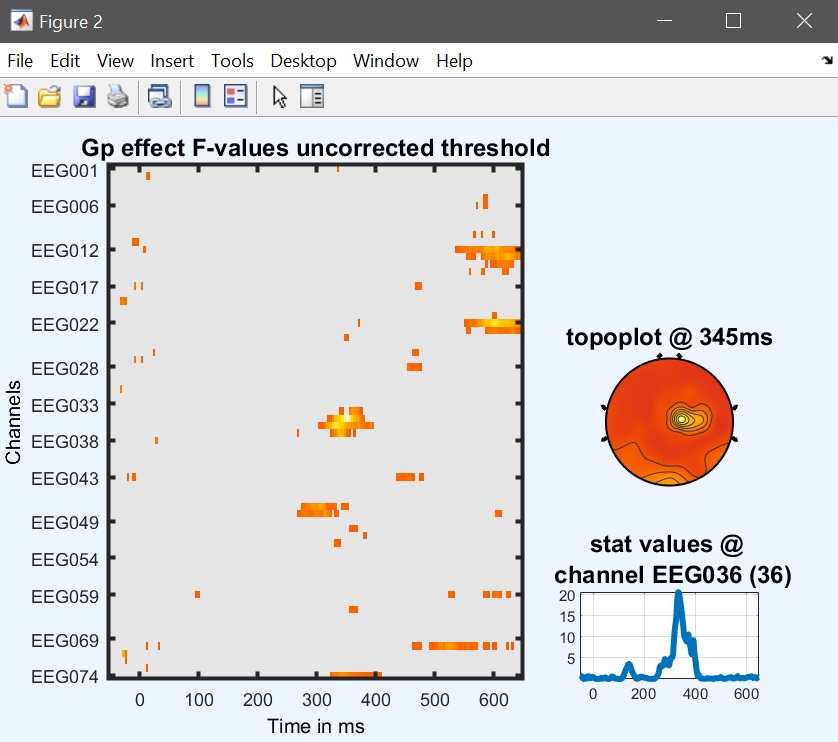
Figure 37a. Gp results ERP
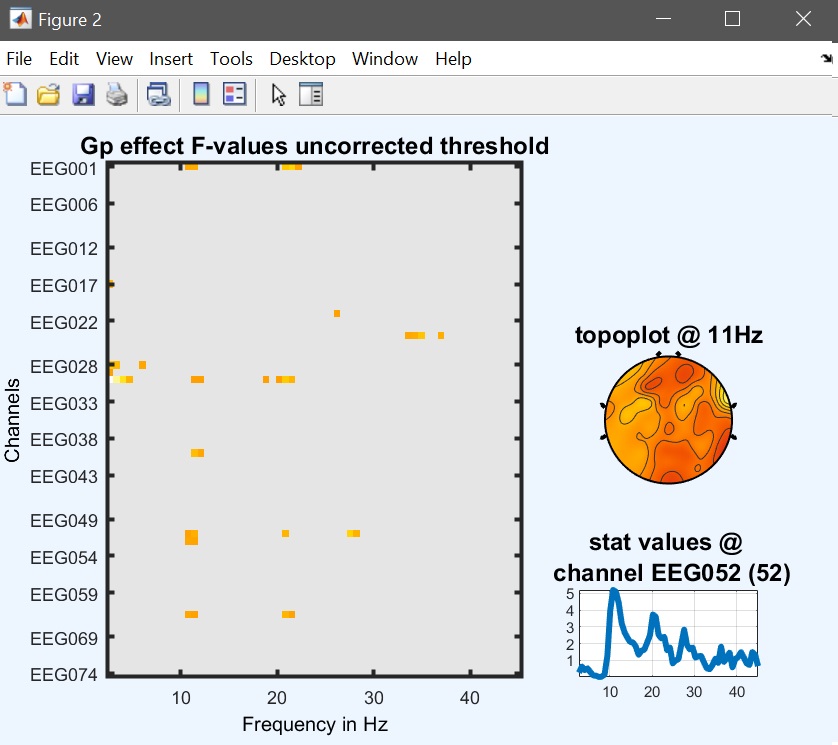
Figure 37b. Gp results Spectrum
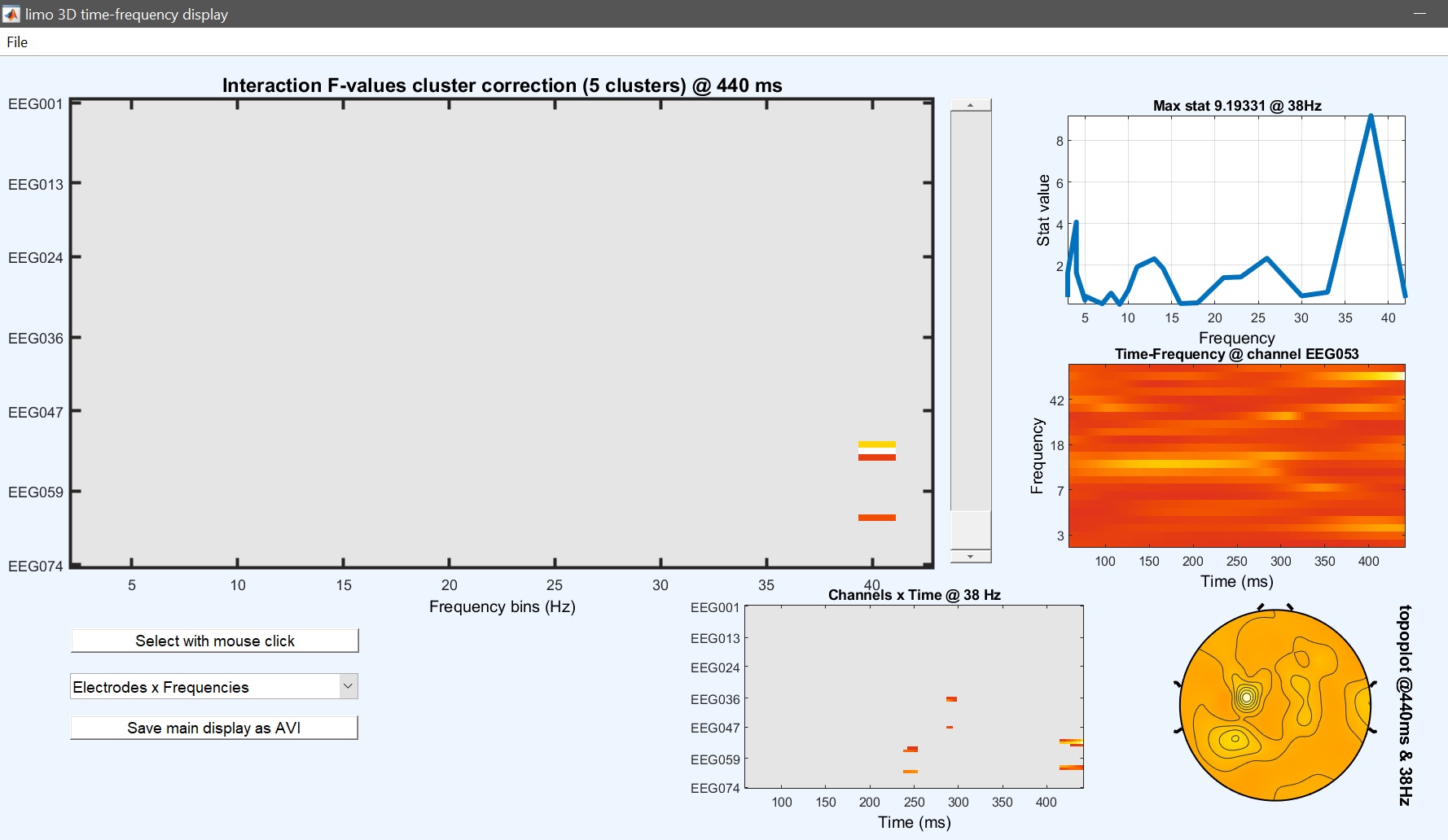
_Figure 37c. GpFace interaction results ERSP_