Importing and managing event and epoch information
This tutorial describes importing, modifying, selecting, and visualizing EEGLAB events within the EEGLAB graphic interface.
EEGLAB counts records of the time and nature of experimental events to analyze the EEG data. This section details how to load in events’ information embedded in one of the data channels, stored in a MATLAB array, or separate ASCII file. Once event information is imported, EEGLAB copies the resulting EEG.event structure to a back-up (ur) copy, EEG.urevent, and creates links from each event to the corresponding urevent. This allows the user to select events based on the previous (or future) event context, even after data containing some events has been rejected from the data, as described in the event scripting section of the tutorial.
Table of contents
- Importing events
- Managing events
Importing events
Events can be imported into EEGLAB by selecting the File → Import event info menu item. Different methods to import events are detailed below.
Importing events from a data channel
Information about experimental events is often recorded onto one of the rows (channels) of the EEG data matrix. Once more, we create simulated data to illustrate how to import events from a data channel. Assuming an EEG dataset with 33 rows (channels), out of which the first 32 are data channels, and the last (33) is an event channel with values 1 (stimulus onset), 2 (subject response), and 0 (other). MATLAB code for generating such data follows (to test, copy and paste the code to the MATLAB command line):
eeglab
eegdata = rand(32, 256*100); % 32 channels of random activity (100 s sampled at 256 Hz).
eegdata(33,[10:256:256*100]) = 1; % simulating a stimulus onset every second
eegdata(33,[100:256:256*100]+round(rand*128)) = 2; % simulating reaction times about 500 ms after stimulus onsets
After copying the code above to MATLAB and importing the array eegdata into EEGLAB as a test dataset using the File → Import data → from ASCII/float file or MATLAB array menu item, select File → Import event info → from data channel menu item to call function pop_chanevent.m .
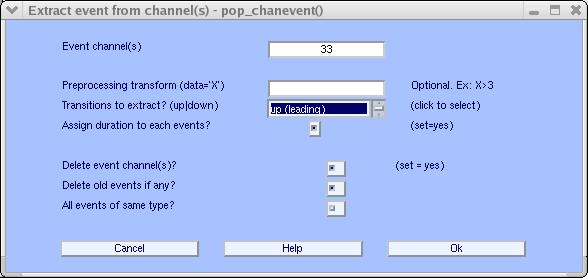
Enter 33 as the event channel and set the edge-extract type to up (leading) (Note: place the mouse over the text Transitions to extract to see contextual help messages).
Press Ok. Now, the event information will have been imported into the test EEGLAB dataset. At the same time, channel 33 will have been deleted from the test data. Select menu item Edit → Event values to inspect the imported event types and latencies.
Importing events from a MATLAB array or text file
Using the random EEG dataset created above, we import event information stored in an ASCII text file, tutorial_eventtable.txt. This text file is composed of three columns, the first containing the latency of the event (in seconds), the second the type of the event, and the third a parameter describing the event (for example, the stimulus position). For example, the top lines of such a file might be:
| Onset | Type | Position |
| 1 | target | 1 |
| 2.3047 | response | 1 |
| 3 | target | 2 |
| 4.7707 | response | 2 |
| 5 | target | 1 |
| 6.5979 | response | 1 |
| ... | ... | ... |
Select menu item File → Import event info → Import MATLAB array or ASCII file
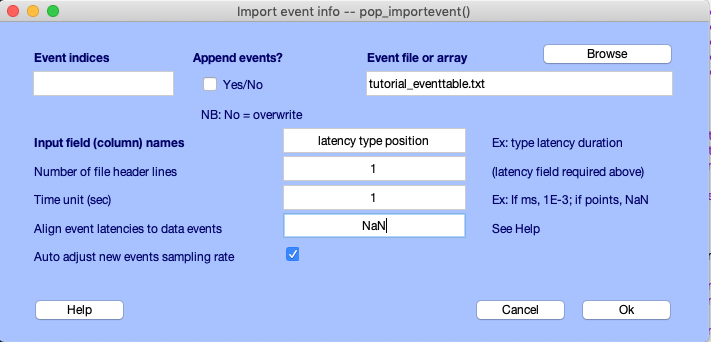
Browse for the tutorial text file tutorial_eventtable.txt, set the number of header lines to 1 (for the first line of the file, which gives the column field names), and set the input fields (i.e., the names associated with the columns in the array) to latency type position. If these field names are quoted or separated by commas, these extra characters are ignored. (NOTE: It is necessary to use the names latency and type for two of the fields. EEGLAB uses these two field names to extract, sort, and display events. These fields must be lowercase since MATLAB is case sensitive.) Event latencies may be in seconds, milliseconds, or samples in the text file, although they are always stored in EEGLAB in samples. In this interactive window, the input Event indices and checkbox Append events? may be used to insert new events instead of replacing, which is the default behavior, a subset of events with new events (for instance, for large EEG files, which may have several event files).
Important note about aligning events
An essential input above is Align event latencies to data events, which aligns the first event latency to the existing event latency and checks latency consistency. A value of NaN (MATLAB for not-a-number) indicates that this option is ignored (as in the example above). However, for most EEG data, the EEG is recorded with basic events stored in an event channel (see Import events from a data channel above), for instance. Detailed event information is recorded separately in a text file: as a consequence, the events recorded in the text file have to be aligned with the events recorded in the EEG.
To do so, set the input for Align event latencies to data events to 0 if the first event in the text file corresponds to the first event recorded in the EEG (i.e. if the offset between the two is 0). Setting this value to 1 indicates that event 1 in the event text file corresponds to event number 2 in the EEG data. Here, negative values can also be used to indicate that events in the text file start before those recorded in the EEG).
When aligning events, as shown in the following section, the function displays the latencies of the two event types, so the user can check that they are aligned based on his knowledge of the experiment (for instance, there may be more events in the text file than recorded in the EEG). The last checkbox automatically adjusts the new events’ sampling rate, so they best align with the closest old events. This may correct small differences in sampling rates that could lead to significant differences by the end of the experiment (e.g., a 0.01% clock difference would lead to a 360-ms difference after one hour if not corrected).
Importing events from a Presentation file
EEGLAB can also import events saved using the Presentation software. Sample files are available for download here: TEST.SMA (a SnapMaster .SMA file) and TEST.LOG (a Presentation event log file). If you click on the links and a text file appear, right click on the link and use Save link as to save the files.
To test the use of these sample files, first import the .SMA data file using menu item File → Import data → From .SMA data file. Then select File → Import event info → from Presentation LOG. file to import events from the Presentation log file as shown below</span>
Then the following window pops-up</span>
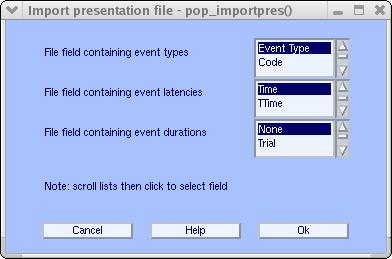
Scroll file fields to select which field (i.e., file column) contains the event type and which column contains the event latency. The default value is fine for this specific file, so simply press Ok. MATLAB then returns:
Replacing field 'Event Type' by 'type' for EEGLAB compatibility
Replacing field 'Time' by 'latency' for EEGLAB compatibility
Renaming second 'Uncertainty' field
Reading file (lines): 6
Check alignment between pre-existing (old) and loaded event latencies:
Old event latencies (10 first): 10789 21315 31375 41902 51962 62489 ...
New event latencies (10 first): 10789 21315 31376 41902 51963 62489 ...
Best sampling rate ratio found is 0.9999895. Below latencies after adjustment
Old event latencies (10 first): 10789 21315 31376 41902 51963 62488 ...
New event latencies (10 first): 10789 21315 31375 41902 51962 62489 ...
Pop_importevent warning: 0/6 have no latency and were removed
eeg_checkset: value format of event field 'Duration' made uniform
eeg_checkset: value format of event field 'Uncertainty2' made uniform
eeg_checkset note: creating the original event table (EEG.urevent)
Done.
The function aligns the first event latency recorded in the Presentation file to the first event latency recorded in the EEG in the SnapMaster file. Check that the events recorded in the SnapMaster file have the same latencies as the ones recorded in the .LOG presentation file. The function then computes the best sampling rate ratio: this may account for small differences in sampling rate that could lead to big differences at the end of the experiment (e.g., 0.01% clock difference during half an hour would lead to a 360-ms difference after one hour if not corrected). Note that if the events are shifted (with respect to events from the binary EEG file), it is always possible to suppress events manually or to import the presentation file as a text file, as described in the previous section. Note that some Presentation files that contain comments at the end of the file may not be supported. If importing a Presentation file fails, try removing any comments from the end of the file. If it still does not work, try importing the Presentation file as a text file, as described in the previous section.
Note: The presentation file contains more events (such as reaction time) than the raw EEG data file, which is why we are importing it. Once events are aligned, the function will automatically remove duplicate events.
Importing E-Prime information files
The E-prime format is highly configurable, so you may use the ASCII importer to import data files. Use the File → Import event info → from E-Prime ASCII (text) file menu item, which is the same as calling the Import data → From ASCII/float file or MATLAB array menu item. Configure the interface with the name of the E-Prime columns to import the event file. In some cases, it might be necessary to export the E-Prime to a tab-delimited file first (for example, in a spreadsheet application) and edit some of the column information that might not be read correctly under MATLAB. Please send us your E-Prime files (at eeglab at sccn.ucsd.edu), so we may tailor the ASCII import menu for E-Prime files.
Importing Neuroscan .DAT information files
A sample .DAT file TEST.DAT associated with the sample TEST.CNT continuous file are available for download for testing purposes. Both the .DAT file and the .CNT files must contain the same number of events (in this case, 100). Neuroscan .DAT files contain data epoch information, so after importing the CNT file into EEGLAB, you must first extract data epochs. To do so, use the menu item Tools → Extract epochs. Select a time window from -0.1 to 0.5, and make sure 100 epochs are generated (if your time window is too large, some epochs at the onset and outset might be removed). Then to import the Neuroscan .DAT file, use the menu item File → Import epoch info → From Neuroscan .DAT info file. After selecting the file to import, a second window appears:
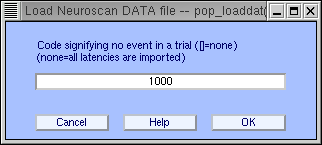
In .DAT files, there must be a reaction time (in milliseconds) for each epoch. However, depending the experiment design, there may be no reaction time in a given epoch. Then one has to use a code value for reaction time latencies in these epochs. For instance, you might decide that a value of 1000 (ms) would indicate that the subject did not respond. (If all the epochs of the experiment already have a reaction time, do not enter anything here.)
Importing epoch info (MATLAB array or text file) into EEGLAB
Importing epoch information means that data epochs have already been extracted from the continuous EEG data and that the MATLAB array or text epoch information file has one entry per epoch. To illustrate how to import such a file or array, we will once more create some simulated EEG data.
eegdata = rand(32, 256, 10); % 32 channels, 256 time points per epoch, 10 epochs
Select menu item File → Import data → From ascii/float data file or MATLAB array. Refer to the previous section of the tutorial.
The MATLAB array, being 3-D, is automatically imported as data epochs: the first dimension is interpreted as data channels, the second as data points, and the third as data epochs or trials (e.g., our sample data matrix above contains 10 epochs). Let us imagine that our simulated EEG data came from a simple stimulus/response task, with subject responses being either ‘correct’ or ‘wrong’ and response latencies recorded in milliseconds. Then the epoch event file might look something like this:
| Epoch | Response | Response_latency |
| 1 | Correct | 502 |
| 2 | Correct | 477 |
| 3 | Correct | 553 |
| 4 | Correct | 612 |
| 5 | Wrong | 430 |
| ... | ... | ... |
This file tutorial_epoch.txt may be downloaded (or copied from the array above in a text file). Then select menu item File → Import epoch info → from MATLAB array or ascii file, bringing up the following window:
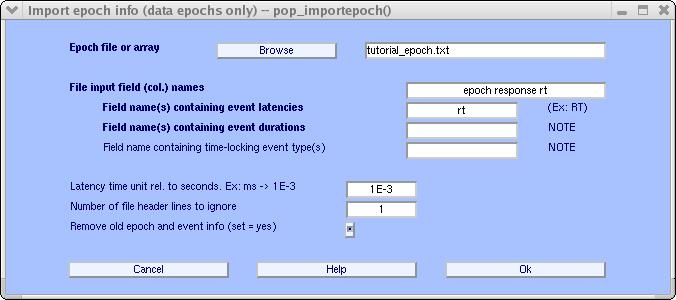
Above, browse for the tutorial_epoch.txt file, set the input fields to epoch response rt (where rt is an acronym for ‘reaction time’). The only one of these fields that contains latency information is rt, so it is entered as input to the Field name(s) containing latencies query box. This file (see above) has 1 header line, as we need to specify in the Number of file header lines to ignore box. Finally, the reaction times are recorded in milliseconds, which we indicate as 1E-3 (i.e., one-thousandth of a second). Note that the last entry, Remove old epoch …, allows the user to import other information later if it is unset. Press Ok, when done. Now select the Edit → Event values menu item to inspect what happened to the reaction time information (use the arrow to move to the second event):
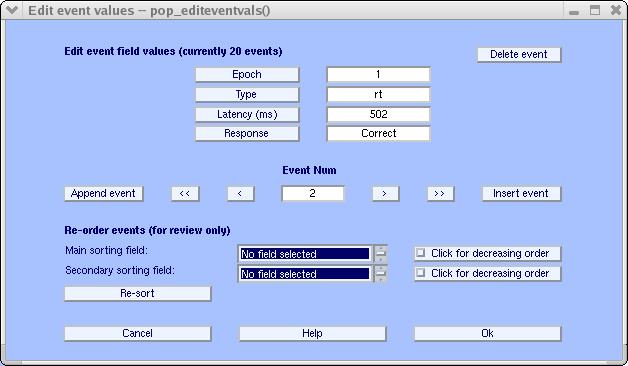
As shown above, when epoch information was imported, events with the type named rt were created and assigned a latency. If we had had several columns containing latency information, the function would have created several types. If you get an error that the number of trials (10, in this case, do not match the number of trials in the EEG dataset), this is because you have not imported the “eegdata” variable properly.
Note: For convenience, standard epoch information is available from the command line in the variable EEG.epoch. Also, event information available in EEG.event can be used for script or command line data processing. See the events script writing tutorials for more details.
Managing events
The current section should help familiarize yourself with event handling in EEGLAB. Most EEGLAB functions take events into account. For instance, functions that remove data (pop_eegplot.m, pop_select.m) will also remove events that occur during the removed data (though not their corresponding urevents). Functions that process continuous data (pop_spectopo.m, pop_resample.m, pop_mergeset.m) will take into account ‘boundary’ events (data events added by EEGLAB to note portions of the data that have been removed or at ‘hard’ boundaries between concatenated datasets). The event type field can be used to extract data epochs, and events can also be used for making ERP-image plots using the pop_erpimage.m function. Finally, event information is extensively used for group-level analysis.
Plotting events
First, start EEGLAB, and load dataset eeglab_data.set in the sample_data folder using the File → Load dataset menu item. Then use the Plot → Channel data (scroll) menu item to visualize the data and associated events.
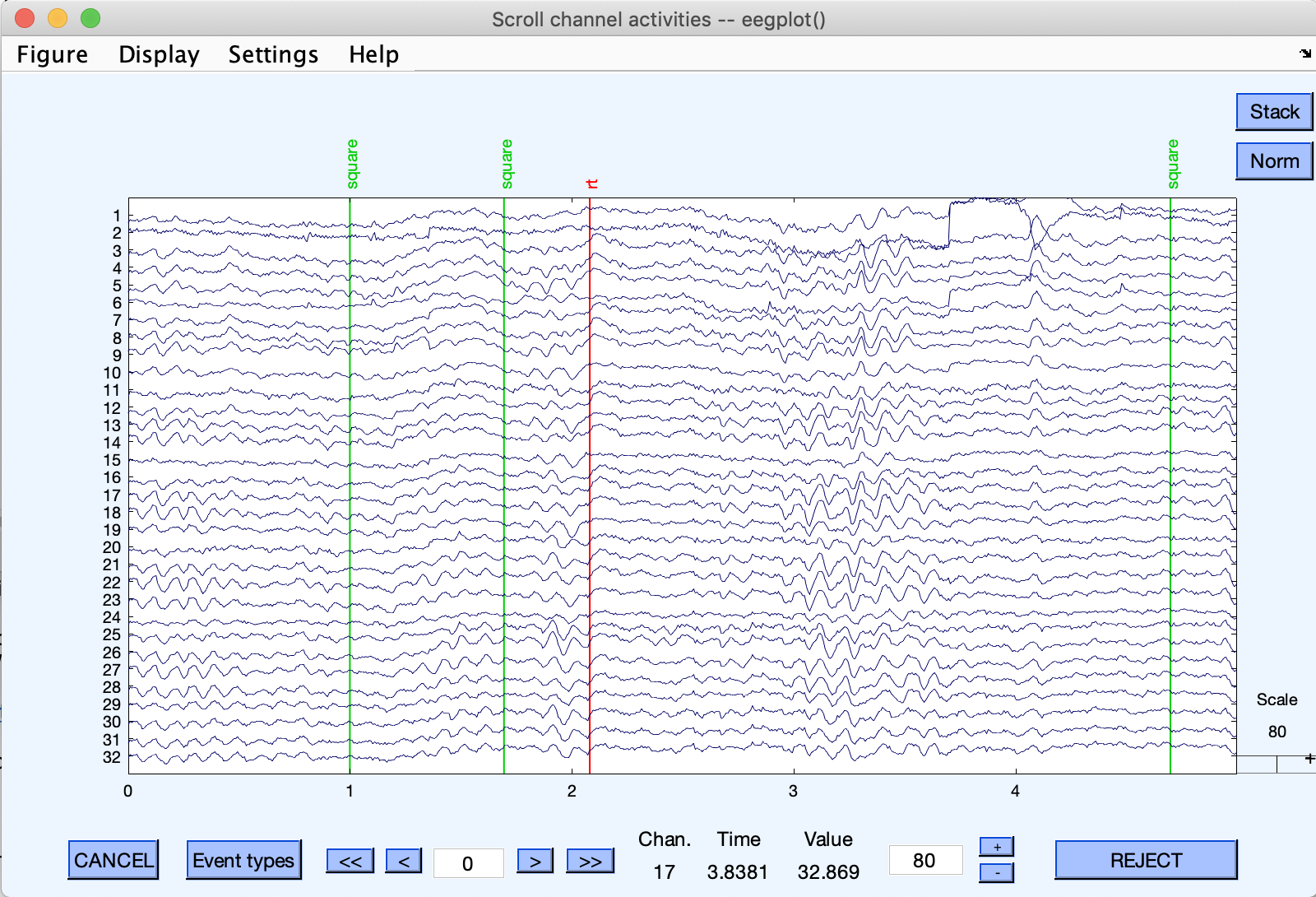
Events are shown as vertical lines. Different event types are shown in different colors. The button Event types will show the list of event types for this dataset.
Recognized event fields
The EEG event structure contains records of the experimental events that occurred while the data was being recorded. To view the information for an event, use the Edit → Event values menu item (see the window below, which shows the tutorial dataset’s events imported in the previous step).
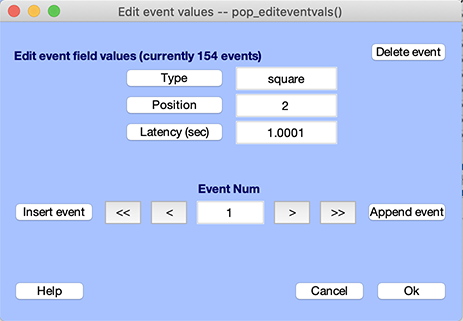
The type and latency fields are the most important EEGLAB event fields (see below). Some EEGLAB functions recognize and use these two fields for plotting events, sorting trials, etc. (Note: One can also process events lacking these fields, though this strongly limits the range of event processing possibilities). Other fields, including epoch, duration, urevent, are also recognized by EEGLAB (they are created automatically during extracting of epochs, rejecting data, or storing event data). User-defined fields can have any other name relevant to the data (for example, position in the example above). A short description of recognized event fields is given below. Further information may be found in the event scripting tutorial.
- type - specifies the type of event. For example, ‘square’ in the example above is a stimulus type, ‘rt’ is a subject button-press (i.e., reaction-time) event, etc… In continuous datasets, EEGLAB may add events of type ‘boundary’ to specify data boundaries (breaks in the continuous data).
- latency - contains event latencies. The latency information is displayed in seconds for continuous data or in milliseconds relative to the epoch’s time-locking event for epoched data. As we will see in the event scripting section, the latency information is stored internally in data samples. These may be fractional samples in case the time resolution of events exceeds the data resolution.
- duration - duration of the event. This information is displayed in seconds for continuous data, and in milliseconds for epoched data. Internally, duration is stored in data samples.
- urevent - contains indices of events in the original (‘ur’ in German) event structure. The first time events are imported, they are copied into a separate structure called the ‘urevent’ structure. This field is hidden in the graphic interface (above) since it should not be casually modified. This field is described in detail in the event scripting tutorial.
- epoch - indices of the data epochs (if any) the event falls within. This field is only present for data for which data epochs have been extracted (which is not the case here since the data is continuous).
Note: all event fields may contain either numbers or strings (except for latency, duration, urevent, and epoch, which must contain numbers). For instance, the type field can contain a number (e.g., 1, 2, etc.) or a string (e.g., ‘square’, ‘rt’, etc.). Note that when using the ERPLAB plugin, the event type field is expected to contain numerical values. EEGLAB cannot process a mixture of both formats (numeric and string) for one field. A function that checks the event structure (eeg_checkset.m with flag eventconsistency), called each time a dataset is modified, enforces each field’s contents to remain consistent, automatically converting numeric values to strings if necessary. This function will also automatically resort events by increasing latencies.
Adding, and modifying events
To insert new events manually, select the Edit → Event values menu item. Click on the Insert event button to add a new event before the current event. The Append event button adds an event after the current event. After a new event has been inserted or appended, its event-field information can immediately be changed in the corresponding boxes. For instance, to insert an event of type new 500 ms after the onset of the time-locking stimulus, enter new in the event type edit box, and 500 in the event latency edit box. Note: If you click on the cancel button, none of the new information will be saved.
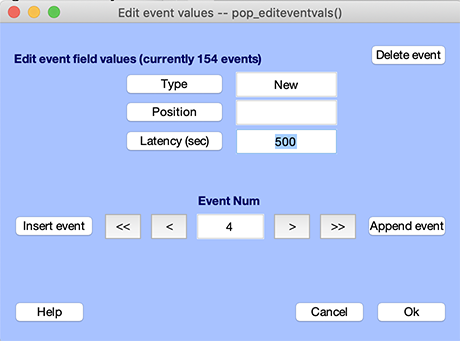
After you press Ok, the events may be resorted (events must always be in order of increasing latencies), and some field contents may be modified to ensure consistency, as indicated at the end of the previous section.
In the graphic interface above, all experimental events can be manually modified by simply typing the new values in the edit window for each field. Events may also be deleted (Delete event button).
The Vised EEGLAB plugin available from the EEGLAB plugin manager allows users to manually add events directly on the EEG browser and is convenient for marking events of interest (such as blinks, for example).
Modifying events in bulk
Another way to modify new events is to export them, modify them under Excel, and then import them back into EEGLAB. You may import the tutorial data eeglab_data.set in the sample_data under EEGLAB, then call menu item File → Export → Events to text file and save a data file. Using this data file, we show how to rename all events with type “rt” to events with type “response” under Excel. The figure below shows the exported file (left) and the edited file (right).
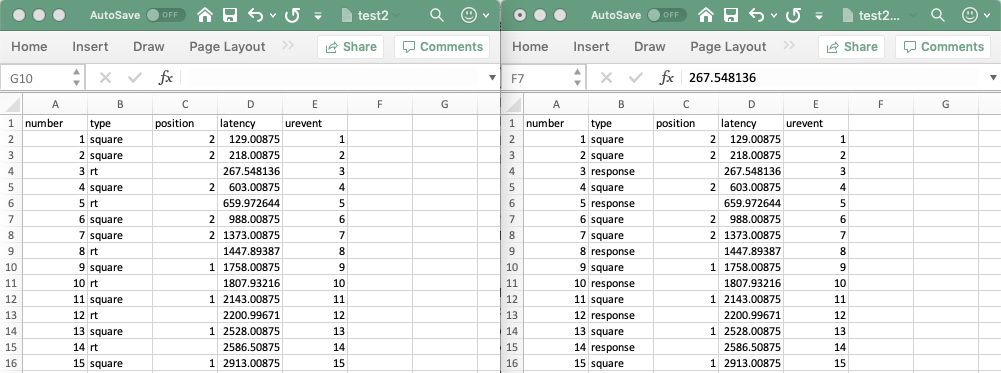
Then, using the File → Import event info → From MATLAB array of ASCII file menu item, we select the modified file. We enter the column names, indicate that there is 1 line of header, and set the unit latency to NaN (which indicates that time information conversion is necessary). We also uncheck the alignment option (although leaving it checked has no effect). Now all the events have been renamed (note that the interface to select events presented in the following section also allows to rename event type values, so this is only presented as a didactic example).
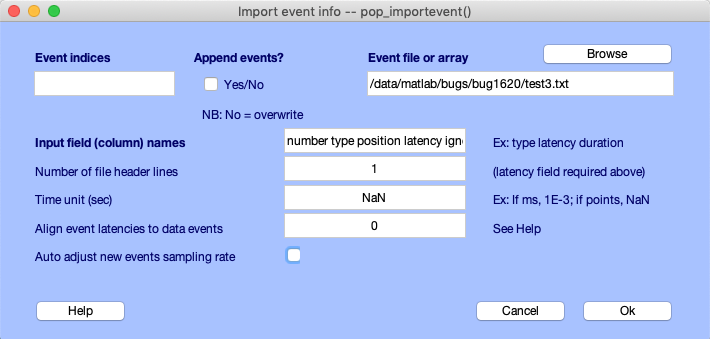
Selecting/removing/renaming events
You may import the tutorial data eeglab_data.set in the sample_data under EEGLAB. To select specific events, use the Edit → Select epochs or eventsmenu item. This can be used to remove spurious events or rename events.
For example, to only keep type “square” events, you would enter “square” for event type as shown below.
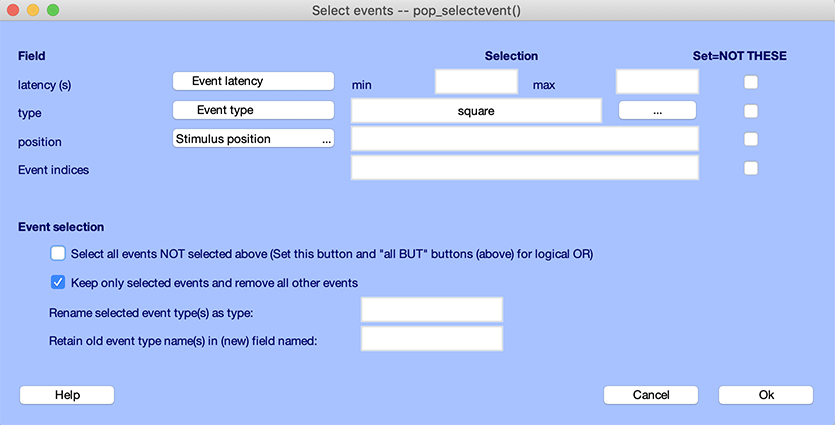
Alternatively, since they are only two event types (“square” and “rt”), you could also remove all reaction time events. To do so, enter “rt” for event type and check the checkbox to select all other events as shown below.
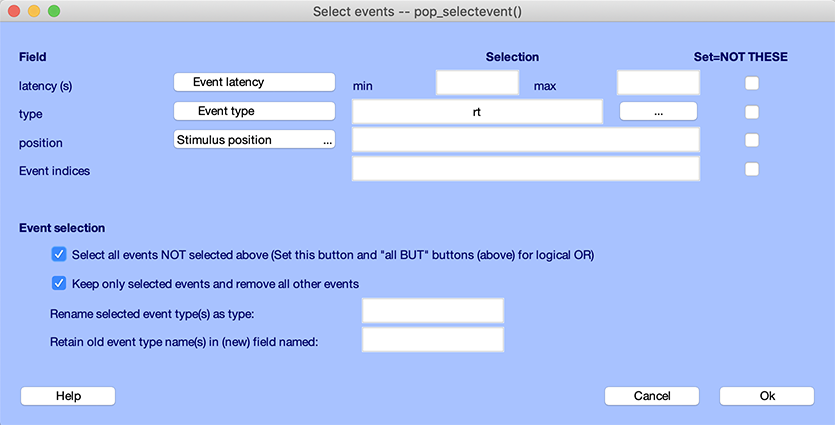
In the example below, we simply rename to “response” events with the type “rt”, as we did in the previous section. Select “rt” for the event type, enter “response” in the renaming event text box. Make sure to uncheck the option Keep only selected events and remove all other events. Otherwise all events which are not of type “rt” will be removed.
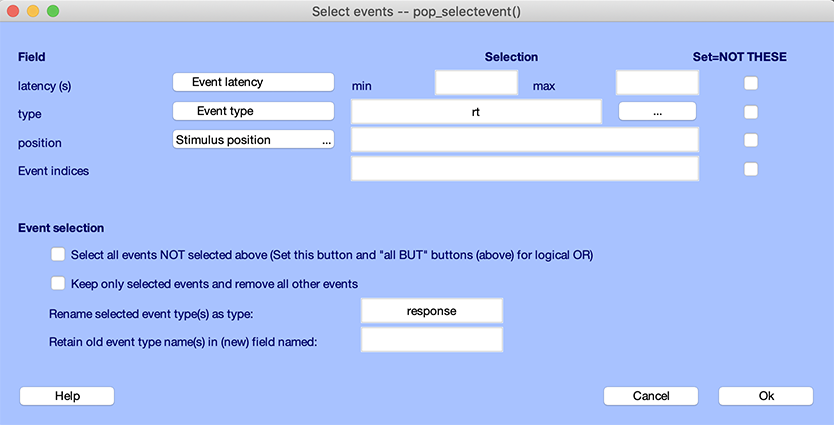
Note: To select events outside the given range, check the Select events NOT selected above box to the right of the field range entry. It is possible to rename the type of the selected events (while (optionally) keeping the old event type name as a new field) using the last edit boxes.
You may also specify more complex combinations of event field selections and ranges as the criterion for selecting trials. For instance, you could select events of type “rt” that have latencies between 0 and 400 milliseconds and rename these as “fast_rt.” Use the event selection interface a second time and rename the remaining events as “slow_rt.”