Let say you only want to know if famous faces differ from unfamiliar faces – again an ANOVA could be set up test the main effect and using contrasts. Alternatively, if that is the only effect of interest, you can compute a contrast at the subject level and do a paired- t-test on contrasts.
Crucially, let’s say the research question is familiar vs unfamiliar and scrambled are just a control - doing the ANOVA is a little meaningless because you now include scrambled as a condition when in fact it’s a control - using contrasts we can compute those differences [1 1 1 -1 -1 -1 0 0 0 0 ; 0 0 0 -1 -1 -1 1 1 1 0] and then compute a paired t-test familiar vs unfamiliar controlled for scrambled. Note however, that the contrast within the ANOVA also controls for the presence of scrambled in the model.
1st level
compute 2 contrasts as described previously, or in command line:
cd(STUDY.filepath)
[~,~,contrast.LIMO_files] = limo_get_files([],[],[],...
fullfile(STUDY.filepath,['LIMO_' STUDY.filename(1:end-6)],...
'LIMO_files_FaceRepAll_GLM_Channels_Time_WLS.txt'));
contrast.mat = [1 1 1 -1 -1 -1 0 0 0 ; 0 0 0 -1 -1 -1 1 1 1];
limo_batch('contrast only',[],contrast);
2nd level
From the 2nd level menu (figure 9), click on ‘paired t-test’, and select the contrasts computed previously for each condition (i.e. con1 and con3). Results (figure 34 for ERP, Spectrum and ERSP) are very similar to figure 33 (ERP, Spectrum and ERSP i.e. the post-hoc test from the 2-way ANOVA and differences relates to the fact that different amount of variance is accounted for, and the paired t-test uses trimmed means across subjects.
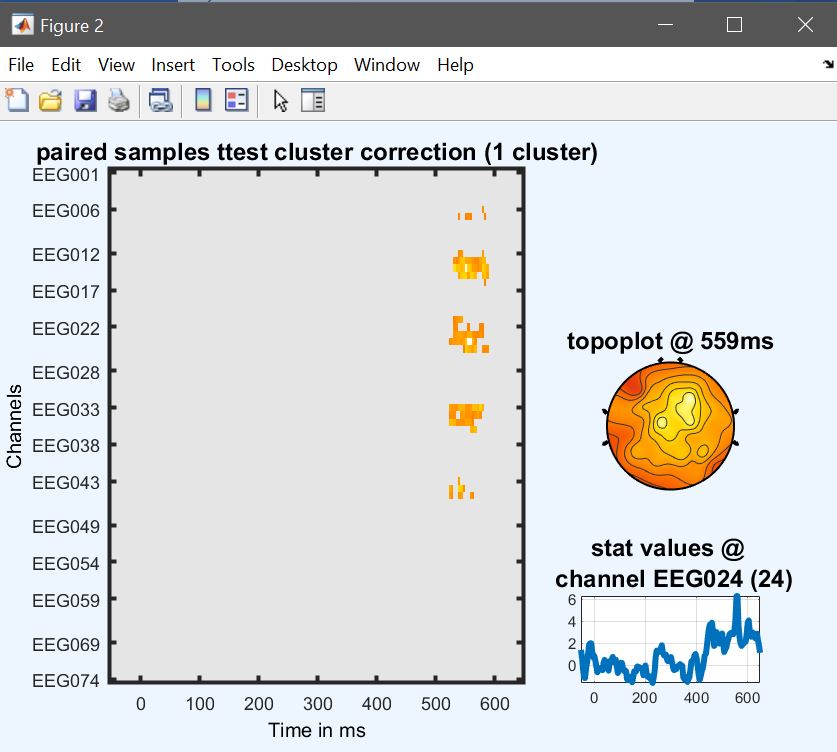
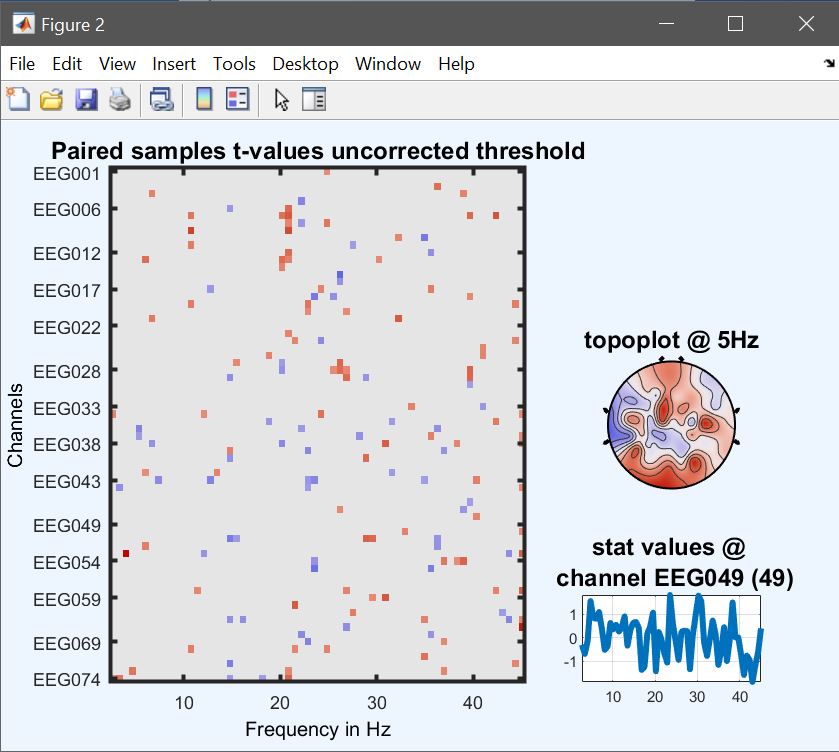
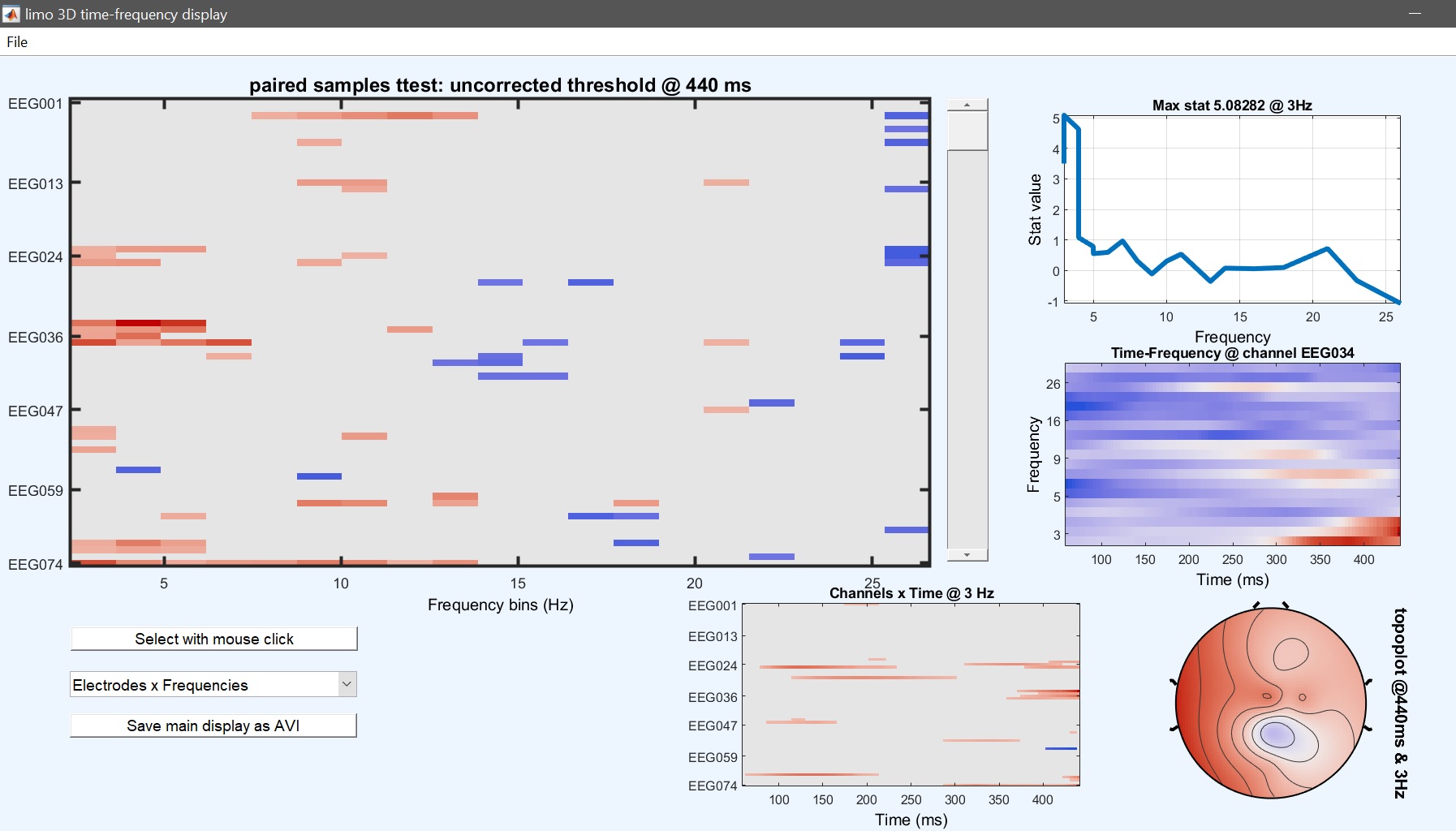
Figure 34. Contrast famous vs unfamiliar
This can be executed in command line as:
mkdir('Paired_ttest'); cd('Paired_ttest');
chanlocs = [STUDY.filepath filesep 'limo_gp_level_chanlocs.mat'];
files = {fullfile(STUDY.filepath,['LIMO_' STUDY.filename(1:end-6)],'con_5_files_FaceRepAll_GLM_Channels_Time_WLS.txt'), ...
fullfile(STUDY.filepath,['LIMO_' STUDY.filename(1:end-6)],'con_6_files_FaceRepAll_GLM_Channels_Time_WLS.txt')};
limo_random_select('paired t-test',chanlocs,'LIMOfiles',files,...
'analysis_type','Full scalp analysis', 'type','Channels','nboot',1000,'tfce',0);